Microvasculature blood flow simulation using HemeLB
eCSE001-010Key Personnel
PI/Co-I: Miguel O. Bernabeu - UCL; Timm Krüger - University of Edinburgh; Peter V. Coveney, James Hetherington, Bruno Silva - UCL
Technical: Mayeul d'Avezac, Owain Kenway, Ian Kirker, Jens Nielsen, Gary Macindoe - UCL
Relevant Documents
eCSE Final Report: Adding a resolved deformable particle model to a highly-parallel blood flow solver for sparse vascular networks
Project summary
HemeLB is an open source, highly-parallel, lattice-Boltzmann (LB) blood flow simulator which is very well suited for the simulation of blood flow in large vascular trees using state-of-the-art supercomputing resources.
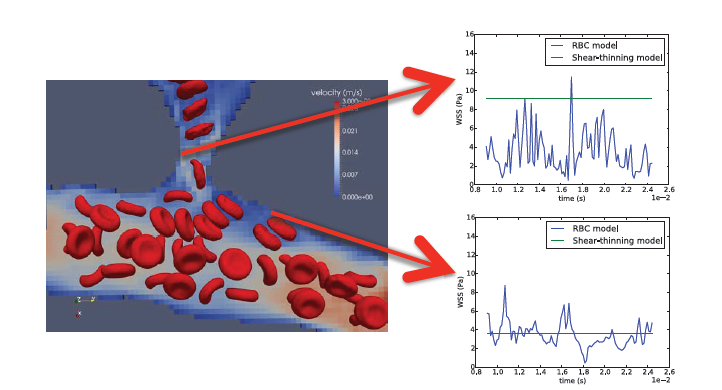
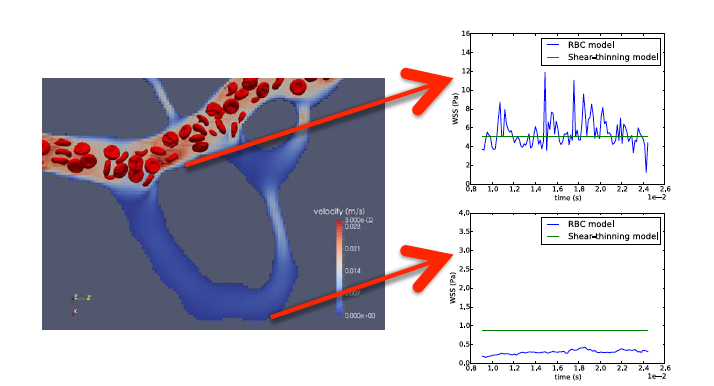
In previous work, HemeLB modelled blood as a shear-thinning homogeneous fluid rather than a complex particle suspension. This simplification is suitable for the study of haemodynamics in larger vessels (i.e. those with diameters larger than a few hundred micrometres). However it fails to capture important haemorheological features when applied to the simulation of blood flow in capillaries, especially as vessel calibre gets closer to, or even smaller than, the typical red blood cell (RBC) diameter of ~8 micrometres. This shortcoming limited the degree of fidelity in the estimates of wall shear stress (WSS).
The main objective of this project was to implement a model of deformable particles into the HemeLB flow solver, to allow the modelling of blood as a dense suspension of deformable particles and facilitate the theoretical study of advanced aspects of haemorheology, oxygen transport, and cell trafficking in realistic vascular networks.
The main components to be implemented were: a) the immersed boundary method for fluid-structure interaction, b) a finite element model of particle membrane elastic deformation, and c) a procedure for simulation initialisation. An important challenge to be overcome in the project was developing a generic implementation of the previous algorithms that could scale up well in terms of domain complexity and particle volume fraction without undermining the excellent parallel scalability of HemeLB.
Thanks to the work done in this eCSE project, we can now model blood as a suspension of deformable particles. The work represents a substantial leap forward for the simulation of blood flow in microvasculature and enables for the first time the theoretical study of advanced aspects of haemorheology, oxygen transport, and cell trafficking in realistic vascular networks. This has already started to provide a much more accurate picture of the WSS experienced by the microvasculature. Furthermore, this will enable the theoretical study of the collective dynamics of dense RBC suspensions in complex vessel geometries.
Summary of the software
The main code used in this project was HemeLB, an open source, highly-parallel, lattice-Boltzmann (LB) blood flow simulator developed by the Centre for Computational Science (CCS), University College London (UCL). HemeLB's main codebase currently receives contributions from additional UK research groups at The University of Edinburgh and Brunel University, London.
HemeLB is a C++/Python application. The main computational kernel is written in C++ and all the pre- and post-processing tools are written in Python. The code is version controlled with Git with a release version being pushed to GitHub (https://github.com/UCL/hemelb). HemeLB is developed using a test-driven approach. Our continuous integration system runs unit and regression tests (currently 136 tests making 448 assertions) after each commit and also every night across a range of supported platforms. Software development is managed using a Scrum process with fortnightly sprints. The HemeLB development team is made of a core of 3-4 developers at UCL and The University of Edinburgh, and a number of external collaborators. HemeLB has successfully run on the following top 500 machines: ARCHER, HECToR, SuperMUC, and Kraken.






