Enabling high performance computing for tools for the analysis of single molecule ion channel currents: probing the energy landscape of channel activation to understand the protein's structure-function relation
eCSE05-06Key Personnel
PI/Co-I: Prof Lucia Sivilotti and Dr James Hetherington - University College London
Technical: Jens Hedegaard Nielsen, Raquel Alegre and Remigijus Lape - University College London
Relevant documents
eCSE Technical Report: HJCFIT ARCHER eCSE end of project technical report
Project summary
HJCFIT is a library for the maximum likelihood fitting of kinetic mechanisms to sequences of open and shut time intervals from single-channel experiments.
Ion channels are involved in vital processes in all living organisms and are the target of many therapeutic drugs. A big step forward in the understanding of ion channel function came when patch clamp experiments made it possible to record the tiny electric current flowing through a single ion channel, as it opens and closes. Long records of such currents contain information about transitions between the discrete conformational states of the channels. The maximum likelihood evaluation involves finding the best estimate for parameters giving the rates of transitions of the channel between kinetic states.
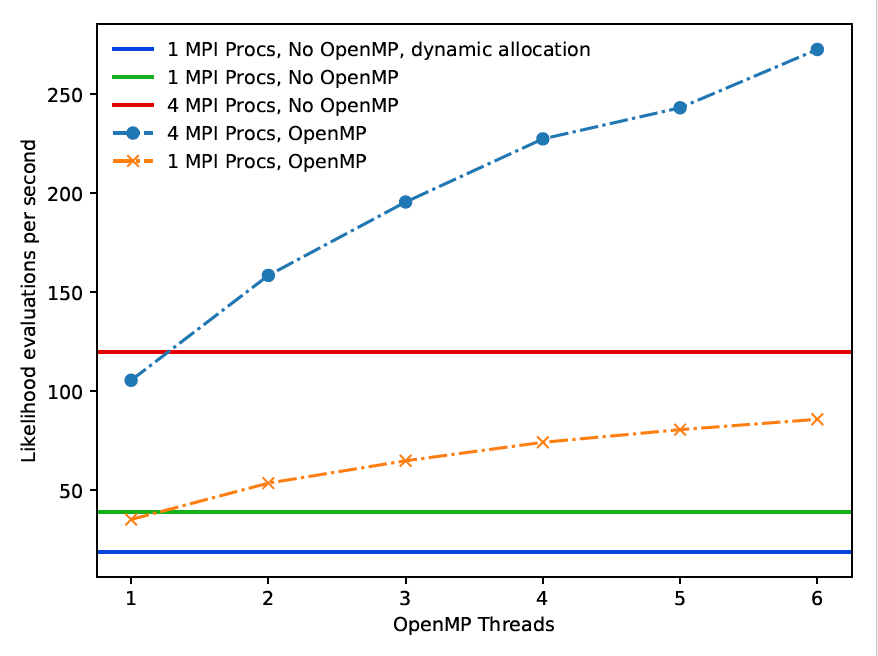
In this project we have transformed HJCFIT from a code suitable to run on individual PCs into a code that runs 14 times faster and is suitable to run on ARCHER (see Figure 1 for examples of the benchmarked performance). Not only does this mean that the time to solution for important scientific problems has been shortened significantly, but it also enables a new community of researchers who have so far not utilised ARCHER or other supercomputing facilities to make use of the code in an efficient way. Firstly, the use of supercomputers enables faster completion of calculations. In some cases, these calculations would have been impossible with the original, non-parallelised code, simpy because the maximum likelihood optimisations would have been too time-consuming to perform. Secondly, on ARCHER, researchers will be able to run many concurrent fitting routines for different experiments or with different parameters. In the past each long calculation job would have totally blocked out a lab computer for a day or longer. Enabling cluster usage we therefore bring this code not only to high performance computing, but also to high throughput computing.
We expect that the speedup through parallelisation will also facilitate and enable new research, making it much more feasible to use the codebase to perform new and interesting calculations based on Bayesian Markov Chain Monte Carlo algorithms. The MCMC methods output is not a single point estimate of a parameter, but its distribution, and thus provides much better information of how accurate and well-defined the rate parameters are. Bayesian methods are in general much more computationally expensive due to the high number of evaluations required.
In addition, we have made the software significantly more robust adding support for the use of multi-precision floating point libraries. Computers in general represent numbers using a finite number of digits. This makes it much more efficient for the computer to do calculations, but sometimes the number of digits used are simply not enough. We could perform all calculations using floating-point numbers with more digits. This would, however, make the code much slower. Instead of that, we have implemented a solution where we perform the calculations using standard floating point numbers and then if that fails we can perform the calculations using higher precision. This gives us the best of both worlds, where the code is more robust without unnecessary sacrifice of speed, as this fallback to the computationally expensive option is performed only in the small set of individual matrix operations where it is needed.
We also added a number of important features to the project documentation such as automatic deployment of the documentation to a number of web pages. Every time the documentation integrated with the code is changed, a webpage version of the documentation is automatically built and deployed to a public facing webpage. The process includes rerunning all the examples of the code and incorporating them in the documentation. An example of the documentation can be seen here: http://dcprogs.github.io/HJCFITdevdocs/.
Achievement of objectives
The first objective was to "re-structure the existing code in order to facilitate maintaining DCProgs in an environment of constant changes in operating systems and hardware platforms". In order to achieve this objective we had to refactor parts of the DCProgs programming suite that are outside HJCFIT (the main part of DCProgs that this project focused on). We found that in order to implement the changes stated in the work-packages, relatively small changes were needed to the rest of the DCProgs suite. We refactored and changed some features in DCProgs to make the package easier to install and support, and to allow it to generate synthetic data for use in DCProgs performance benchmarks.
The second objective was to "enable the use of the ARCHER high-performance computing platform". Once this was achieved, the improvement in performance was to be measured by producing scaling graphs of the maximum likelihood fitting and the Markov Chain Monte Carlo (MCMC) fitting packages. Scaling graphs for maximum likelihood fitting showed a total speedup of 14-fold, which was achieved by a combination of serial optimisations to memory allocation and parallelisation.
The third objective was to "enhance the robustness of the maximum likelihood calculations". The success metrics for this objective applied the calculations to known failure cases. We have identified a particular calculation which represents the typical numerical errors seen. As part of the likelihood calculation HJCFIT needs to calculate the eigenvalues of a matrix, but this calculation fails frequently because the matrix is almost singular. In order to solve this problem, we have incorporated support for multi-precision floating point arithmetic using the standard libraries MPFR and GMP. If needed, the code can still be compiled without this multi-precision support for platforms that do not support it. We have also implemented a transparent switchover, where the code first calculates the eigenvalues using standard double precision. If that fails, the code falls back to the slower multi-precision version. We have shown that this enables calculations that would have failed in the original code.
The fourth objective was to extend our analysis toolkit so it may address new scientific questions in the future. The actual implementation of such new scientific tasks was out of the scope of the present application. This objective has been achieved, because the significant speedup (14 times faster), and the increased robustness of the calculations we produced, open up many new possibilities, where the likelihood can be calculated for systems that are significantly more computationally expensive to evaluate (eg Bayesian inference using MCMC).
Summary of the Software
HJCFIT is a library for maximum likelihood fitting of kinetic models to single-channel current intervals. The HJCFIT outputs the estimates for transition rates between kinetic states. The likelihood for a sequence of open and shut intervals is calculated based on the exact solution of the missed events problem (Hawkes, Jalali and Colquhoun 1990, 1992).
The HJCFIT core kernel is written in C++ with Python bindings. HJCFIT is available as open source licenced under the GPL licence. The code is hosted on Github at https://github.com/dcprogs/hjcfit and developed as an open source project. Most of the changes done as part of this project are available on the develop branch. The rich documentation of the project is atomically published from the master branch at https://dcprogs.github.io/HJCFITmasterdocs/ and from the develop branch at http://dcprogs.github.io/HJCFITdevdocs/. An older but static version of the docs can be found at http://dcprogs.github.io/HJCFIT.
The code is not currently automatically installed on ARCHER but instructions are provided in the documentation along with instructions for other platforms.






